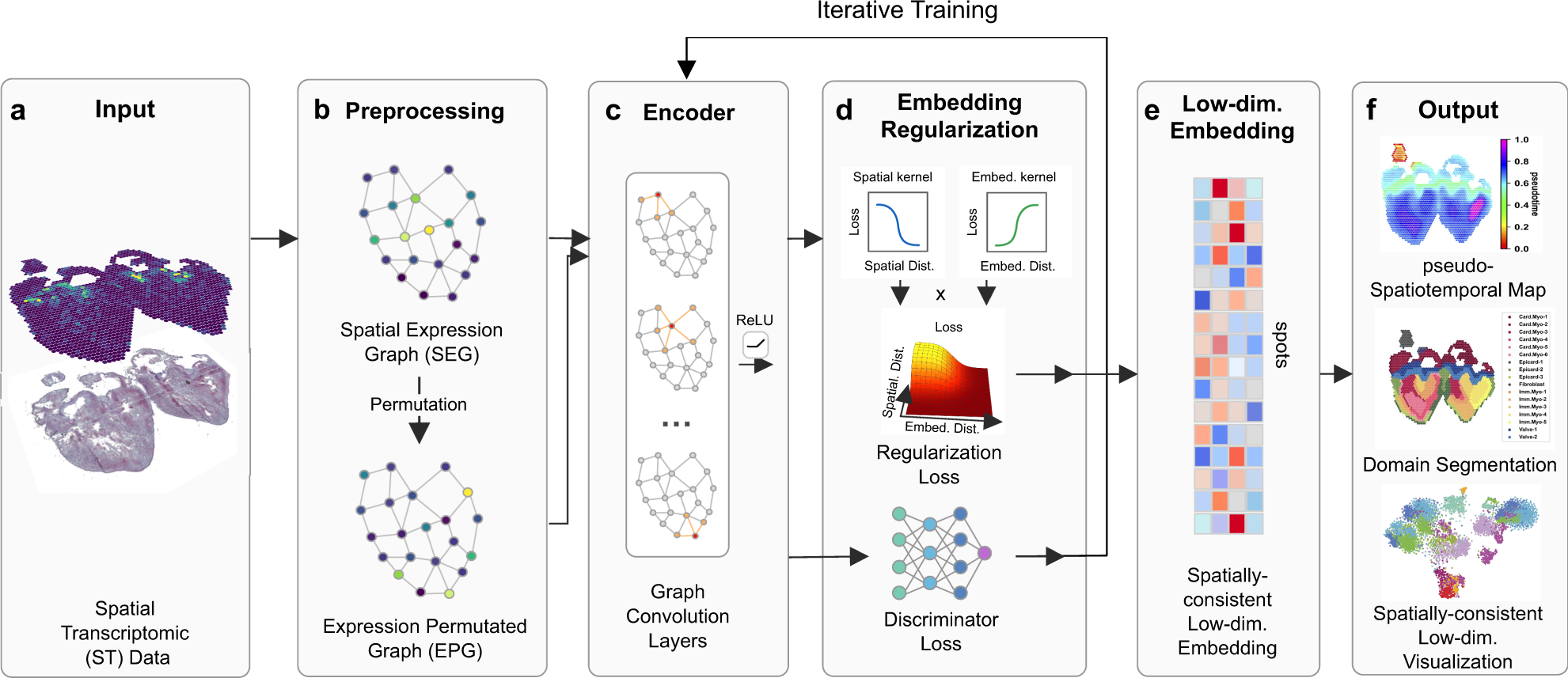
Identifying Pseudo-Spatial Map¶
SpaceFlow is Python package for identifying spatiotemporal patterns and spatial domains from Spatial Transcriptomic (ST) Data. Based on deep graph network, SpaceFlow provides the following functions:
- Encodes the ST data into low-dimensional embeddings that reflecting both expression similarity and the spatial proximity of cells in ST data.
- Incorporates spatiotemporal relationships of cells or spots in ST data through a pseudo-Spatiotemporal Map (pSM) derived from the embeddings.
- Identifies spatial domains with spatially-coherent expression patterns.
Check out (Ren et al., Nature Communications, 2022) for the detailed methods and applications.
import omicverse as ov
#print(f"omicverse version: {ov.__version__}")
import scanpy as sc
#print(f"scanpy version: {sc.__version__}")
ov.utils.ov_plot_set()
____ _ _ __ / __ \____ ___ (_)___| | / /__ _____________ / / / / __ `__ \/ / ___/ | / / _ \/ ___/ ___/ _ \ / /_/ / / / / / / / /__ | |/ / __/ / (__ ) __/ \____/_/ /_/ /_/_/\___/ |___/\___/_/ /____/\___/ Version: 1.6.0, Tutorials: https://omicverse.readthedocs.io/
Preprocess data¶
Here we present our re-analysis of 151676 sample of the dorsolateral prefrontal cortex (DLPFC) dataset. Maynard et al. has manually annotated DLPFC layers and white matter (WM) based on the morphological features and gene markers.
This tutorial demonstrates how to identify spatial domains on 10x Visium data using STAGATE. The processed data are available at https://github.com/LieberInstitute/spatialLIBD. We downloaded the manual annotation from the spatialLIBD package and provided at https://drive.google.com/drive/folders/10lhz5VY7YfvHrtV40MwaqLmWz56U9eBP?usp=sharing.
adata = sc.read_visium(path='data', count_file='151676_filtered_feature_bc_matrix.h5')
adata.var_names_make_unique()
reading data/151676_filtered_feature_bc_matrix.h5 (0:00:00)
Note
We introduced the spatial special svg calculation module prost in omicverse versions greater than `1.6.0` to replace scanpy's HVGs, if you want to use scanpy's HVGs you can set mode=`scanpy` in `ov.space.svg` or use the following code.
#adata=ov.pp.preprocess(adata,mode='shiftlog|pearson',n_HVGs=3000,target_sum=1e4)
#adata.raw = adata
#adata = adata[:, adata.var.highly_variable_features]
sc.pp.calculate_qc_metrics(adata, inplace=True)
adata = adata[:,adata.var['total_counts']>100]
adata=ov.space.svg(adata,mode='prost',n_svgs=3000,target_sum=1e4,platform="visium",)
adata.raw = adata
adata = adata[:, adata.var.space_variable_features]
adata
Filtering genes ... Calculating image index 1D:
100%|██████████| 3460/3460 [00:00<00:00, 16833.57it/s]
Normalize each geneing... Gaussian filtering...
100%|██████████| 5779/5779 [00:13<00:00, 435.78it/s]
Binary segmentation for each gene:
100%|██████████| 5779/5779 [00:14<00:00, 400.79it/s]
Spliting subregions for each gene:
100%|██████████| 5779/5779 [00:35<00:00, 160.68it/s]
Computing PROST Index for each gene:
100%|██████████| 5779/5779 [00:03<00:00, 1838.06it/s]
PROST Index calculation completed !! PI calculation is done!
100%|██████████| 5779/5779 [03:11<00:00, 30.22it/s]
Spatial autocorrelation test is done!
normalizing counts per cell
finished (0:00:00)
normalization and log1p are done!
3000 SVGs are selected!
View of AnnData object with n_obs × n_vars = 3460 × 3000
obs: 'in_tissue', 'array_row', 'array_col', 'n_genes_by_counts', 'log1p_n_genes_by_counts', 'total_counts', 'log1p_total_counts', 'pct_counts_in_top_50_genes', 'pct_counts_in_top_100_genes', 'pct_counts_in_top_200_genes', 'pct_counts_in_top_500_genes', 'image_idx_1d'
var: 'gene_ids', 'feature_types', 'genome', 'n_cells_by_counts', 'mean_counts', 'log1p_mean_counts', 'pct_dropout_by_counts', 'total_counts', 'log1p_total_counts', 'n_cells', 'SEP', 'SIG', 'PI', 'Moran_I', 'Geary_C', 'p_norm', 'p_rand', 'fdr_norm', 'fdr_rand', 'selected'
uns: 'spatial', 'grid_size', 'locates', 'nor_counts', 'gau_fea', 'binary_image', 'subregions', 'del_index', 'log1p'
obsm: 'spatial'
layers: 'counts'
We read the ground truth area of our spatial data
# read the annotation
import pandas as pd
import os
Ann_df = pd.read_csv(os.path.join('data', '151676_truth.txt'), sep='\t', header=None, index_col=0)
Ann_df.columns = ['Ground Truth']
adata.obs['Ground Truth'] = Ann_df.loc[adata.obs_names, 'Ground Truth']
sc.pl.spatial(adata, img_key="hires", color=["Ground Truth"])
Training the SpaceFlow Model¶
Here, we used ov.space.pySpaceFlow to construct a SpaceFlow Object and train the model.
We need to store the space location info in adata.obsm['spatial']
sf_obj=ov.space.pySpaceFlow(adata)
We then train a spatially regularized deep graph network model to learn a low-dimensional embedding that reflecting both expression similarity and the spatial proximity of cells in ST data.
Parameters:
spatial_regularization_strength: the strength of spatial regularization, the larger the more of the spatial coherence in the identified spatial domains and spatiotemporal patterns. (default: 0.1)z_dim: the target size of the learned embedding. (default: 50)lr: learning rate for optimizing the model. (default: 1e-3)epochs: the max number of the epochs for model training. (default: 1000)max_patience: the max number of the epoch for waiting the loss decreasing. If loss does not decrease for epochs larger than this threshold, the learning will stop, and the model with the parameters that shows the minimal loss are kept as the best model. (default: 50)min_stop: the earliest epoch the learning can stop if no decrease in loss for epochs larger than themax_patience. (default: 100)random_seed: the random seed set to the random generators of therandom,numpy,torchpackages. (default: 42)gpu: the index of the Nvidia GPU, if no GPU, the model will be trained via CPU, which is slower than the GPU training time. (default: 0)regularization_acceleration: whether or not accelerate the calculation of regularization loss using edge subsetting strategy (default: True)edge_subset_sz: the edge subset size for regularization acceleration (default: 1000000)
sf_obj.train(spatial_regularization_strength=0.1,
z_dim=50, lr=1e-3, epochs=1000,
max_patience=50, min_stop=100,
random_seed=42, gpu=0,
regularization_acceleration=True, edge_subset_sz=1000000)
66%|██████▌ | 655/1000 [00:21<00:11, 31.03it/s]
array([[-0.1300505 , 0.50017 , 5.131185 , ..., 9.43627 ,
-0.02699862, -0.6617227 ],
[-0.08247382, 0.46351513, 4.881969 , ..., 8.106182 ,
-0.14856927, -0.3372539 ],
[ 4.2177234 , -1.1615736 , 4.5954304 , ..., 7.9274936 ,
-0.6848111 , -0.42961222],
...,
[ 3.9101906 , -0.9978265 , 4.8047457 , ..., 7.6062427 ,
-0.62523276, -0.37077528],
[ 2.0904627 , -0.27526417, 4.471681 , ..., 6.9050336 ,
-0.5009857 , 0.02095482],
[-0.0645832 , 0.4021829 , 4.3925695 , ..., 7.70629 ,
-0.13314559, -0.29742143]], dtype=float32)
Calculated the Pseudo-Spatial Map¶
Unlike the original SpaceFlow, we only need to use the cal_PSM function when calling SpaceFlow in omicverse to compute the pSM.
sf_obj.cal_pSM(n_neighbors=20,resolution=1,
max_cell_for_subsampling=5000,psm_key='pSM_spaceflow')
computing neighbors
finished: added to `.uns['neighbors']`
`.obsp['distances']`, distances for each pair of neighbors
`.obsp['connectivities']`, weighted adjacency matrix (0:00:04)
computing UMAP
finished: added
'X_umap', UMAP coordinates (adata.obsm) (0:00:04)
running Leiden clustering
finished: found 18 clusters and added
'leiden', the cluster labels (adata.obs, categorical) (0:00:00)
running PAGA
finished: added
'paga/connectivities', connectivities adjacency (adata.uns)
'paga/connectivities_tree', connectivities subtree (adata.uns) (0:00:00)
computing Diffusion Maps using n_comps=15(=n_dcs)
computing transitions
finished (0:00:00)
eigenvalues of transition matrix
[1. 0.99924356 0.9966658 0.99514526 0.9919292 0.9897605
0.98541844 0.9827503 0.97917855 0.97792786 0.97322786 0.9706052
0.96208817 0.96147996 0.9581108 ]
finished: added
'X_diffmap', diffmap coordinates (adata.obsm)
'diffmap_evals', eigenvalues of transition matrix (adata.uns) (0:00:00)
computing Diffusion Pseudotime using n_dcs=10
finished: added
'dpt_pseudotime', the pseudotime (adata.obs) (0:00:00)
The pseudo-spatial map values are stored in adata.obs["pSM_spaceflow"].
array([0.9162037 , 0.8701397 , 0.05179406, ..., 0.06386783, 0.39298058,
0.8717118 ], dtype=float32)
Clustering the space¶
We can use GMM, leiden or louvain to cluster the space.
sc.pp.neighbors(adata, n_neighbors=15, n_pcs=50,
use_rep='spaceflow')
ov.utils.cluster(adata,use_rep='spaceflow',method='louvain',resolution=1)
ov.utils.cluster(adata,use_rep='spaceflow',method='leiden',resolution=1)
ov.utils.cluster(adata,use_rep='spaceflow',method='GMM',n_components=7,covariance_type='full',
tol=1e-9, max_iter=1000, random_state=3607)
running GaussianMixture clustering
finished: found 7 clusters and added
'gmm_cluster', the cluster labels (adata.obs, categorical)