单细胞样本对齐: 模型准备
我们在准备好需要对齐的两个单细胞数据后,便需要开始构建模型了
1. 数据清洗
1.1 导入包
import anndata
import networkx as nx
import scanpy as sc
import scglue
import pandas as pd
import numpy as np
from matplotlib import rcParams
1.2 导入数据
rna = anndata.read_h5ad("/content/GSE174367rna_61472.h5ad")
rna
AnnData object with n_obs × n_vars = 61472 × 58721 obs: 'cell_type', 'Sample.ID', 'Batch', 'Age', 'Sex', 'PMI', 'Tangle.Stage', 'Plaque.Stage', 'Diagnosis', 'RIN' var: 'gene_ids', 'feature_types', 'genome'
atac = anndata.read_h5ad("/content/GSE174367atac_61472.h5ad")
atac
AnnData object with n_obs × n_vars = 61472 × 217707 obs: 'cell_type', 'ran', 'Sample.ID', 'Batch', 'Age', 'Sex', 'PMI', 'Tangle.Stage', 'Plaque.Stage', 'Diagnosis', 'RIN' var: 'feature_types', 'genome', 'chrom', 'chromStart', 'chromEnd', 'n_counts'
1.3 rna数据处理
rna.layers["raw"] = rna.X.copy()
rna.layers["raw"] = rna.X.copy()
rna.var_names_make_unique()
sc.pp.filter_genes(rna, min_cells=3)
sc.pp.normalize_total(rna, target_sum=1e4)
sc.pp.log1p(rna)
sc.pp.highly_variable_genes(rna, min_mean=0.0125, max_mean=3, min_disp=0.5)
1.4 rna数据可视化
sc.tl.pca(rna, n_comps=100, svd_solver="auto")
sc.pp.neighbors(rna, metric="cosine")
sc.tl.umap(rna)
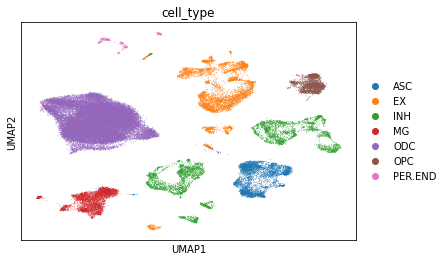
sc.pl.umap(rna, color="cell_type")
rna.write_h5ad('GSE174367rna_61472_process.h5ad',compression="gzip")
1.5 atac数据处理
scglue.data.lsi(atac, n_components=100, n_iter=15)
sc.pp.neighbors(atac, use_rep="X_lsi", metric="cosine")
sc.tl.umap(atac)
sc.pl.umap(atac, color="cell_type")
atac.write_h5ad('GSE174367atac_61472_process.h5ad',compression="gzip")

1.6 Construct prior regulatory graph
因为我们得到的rna只有基因的名字,我们还需要知道他们在染色体上的位置,所以在这里,我们用scglue中标注基因的方法
1.6.1 下载gtf标注文件
#人源
wget ftp://ftp.ebi.ac.uk/pub/databases/gencode/Gencode_human/release_31/gencode.v31.chr_patch_hapl_scaff.annotation.gtf.gz
#鼠源
wget ftp://ftp.ebi.ac.uk/pub/databases/gencode/Gencode_mouse/release_M25/gencode.vM25.chr_patch_hapl_scaff.annotation.gtf.gz
1.6.2 标注
scglue.data.get_gene_annotation(
rna, gtf="gencode.v31.chr_patch_hapl_scaff.annotation.gtf.gz",
gtf_by="gene_name"
)
rna.var.loc[:, ["chrom", "chromStart", "chromEnd"]].head()
1.6.3 去除空标注基因
有一些基因并没有在gtf中找到对应的染色体位置,我们需要把这一部分基因去掉
rna.var['dell']=np.zeros(len(rna.var))
a=rna.var[~rna.var['chromStart'].isnull()].index
rna.var.loc[a,'dell']=1
rna1=rna[:,rna.var.dell==1]
rna1.var = rna1.var.astype({"chromStart": int, "chromEnd": int})
rna1
到此,rna与atac数据的预处理已经完成。
2. 多组学网络构建
根据组学层的先验关系,我们用scglue构建分子层之间的关系
2.1 构建图网络
graph = scglue.genomics.rna_anchored_prior_graph(rna1, atac)
graph.number_of_nodes(), graph.number_of_edges()
100%|██████████| 35363/35363 [00:04<00:00, 7923.69it/s]
(253070, 613256)
2.2 检查节点
# Graph node covers all omic features
all(graph.has_node(gene) for gene in rna1.var_names), \
all(graph.has_node(peak) for peak in atac.var_names)
(True, True)
# Edge attributes contain weights and signs
for _, e in zip(range(5), graph.edges):
print(f"{e}: {graph.edges[e]}")
('AL669831.3', 'chr1:629708-630559', 0): {'dist': 0, 'weight': 1.0, 'sign': 1} ('AL669831.3', 'chr1:631640-631948', 0): {'dist': 0, 'weight': 1.0, 'sign': 1} ('AL669831.3', 'chr1:632511-633105', 0): {'dist': 0, 'weight': 1.0, 'sign': 1} ('AL669831.3', 'chr1:633740-634682', 0): {'dist': 0, 'weight': 1.0, 'sign': 1} ('AL669831.3', 'chr1:778234-779324', 0): {'dist': 0, 'weight': 1.0, 'sign': 1}
2.3 保存图网络
rna1.write("rna_preprocessed.h5ad", compression="gzip")
atac.write("atac_preprocessed.h5ad", compression="gzip")
nx.write_graphml(graph, "prior.graphml.gz")
到这里,模型的预处理就算是完成了,更多的可以参照scglue的官方说明文档。